 Pan-cancer genome standards for five clinically
Pan-cancer genome standards for five clinically
actionable variants
The following five clinically-actionable somatic mutations are covered by the International Standards:
- PIK3CA p.E545K
- TP53 p.R306*
- NRAS p.G12C
- PTEN p.K267fs*9
- MAP2K1/MEK1 p.D67N
The standards are comprised of freeze-dried purified genomic DNA (gDNA) extracted from two cancer cell lines, HCT 15 (NIBSC code 18/118) and MOLT-4 (18/130), and the common wild-type cell line ATDB102 (18/164).
HCT 15 is a colon adenocarcinoma cell line, carrying the PIK3CA p.E545K variant. MOLT-4 is an acute T lymphoblastic leukaemia cell line, carrying the TP53 p.R306*, NRAS p.G12C, PTEN p.K267fs*9, and MAP2K1/MEK1 p.D67N variants. ATDB102 is an Epstein-Barr Virus-transformed lymphoblastoid wild-type cell line.
The International Standards can be diluted, either with wild-type material 18/164 or another wild-type gDNA aligned to 18/164, by applying a calculation specific to each standard, to produce standards at a range of variant percentages which enable the calibration of quantitative assays.
An extensive international collaborative study
The International Standards were approved by the WHO based on the results of an extensive collaborative study involving 35 laboratories across 22 countries and 5 continents.
The study aimed to evaluate the suitability of the materials for use as international standards in multiple assays, and to assign a percentage value for each variant.
Accordingly, a consensus variant percentage for each of the five variants has been derived from the average values of 38 different NGS analysis and dPCR methods.
Furthermore, this collaborative work has also identified additional variants present in the three gDNA materials. This information is useful for the broader validation of NGS pipelines, although it is not intended for diagnostic purposes.
Dr Susie Cooke, Head of Medical Genomics at the Glasgow Precision Oncology Laboratory which participated in this study comments that:
“Cancer genomics is rapidly becoming a key part of the diagnostic, prognostic and treatment planning pathways for cancer patients. It is therefore essential that high-quality reference materials are available to ensure delivery of consistent and accurate test results for all patients. Working with NIBSC on this extensive collaborative study has been a valuable experience for my lab and has produced a valuable resource for the cancer genomics community.”
Similarly, our collaborators from the Genomics England Ltd, add that:
“We established that the genomic DNA size fractionation profiles of these materials is compatible with long read sequencing approaches and we are currently characterising these standards to support our R&D activities.”
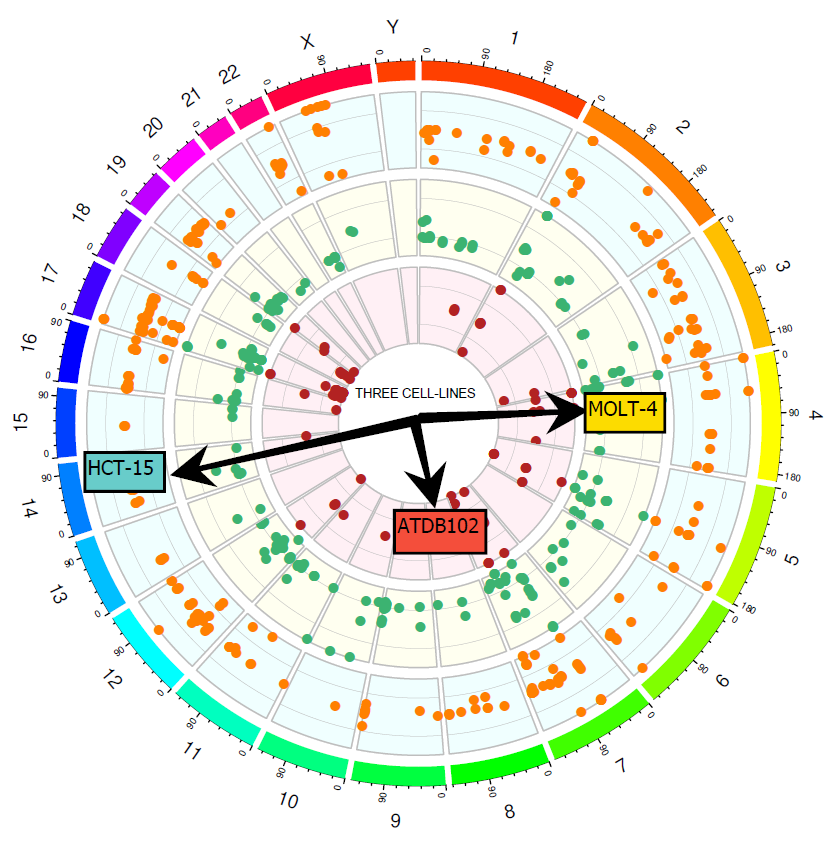
Circos plot showing genomic data for the three International Standards. Each dot represents variants found in the collaborative study for each of the three gDNA, using a wide range of panels with varying coverage.
Part of an ongoing strategy
This work represents part of an ongoing strategy to develop International Standards for cancer genomes. These standards are intended to be used by clinical laboratories worldwide as well as developers of reference materials, kits and assays used for the calibration of actionable variants and broader validation of any NGS pipeline.
The next step in this strategy is to publish a paper detailing the process involved in cancer genome standard development. Following publication, the additional novel variant data will be submitted to the COSMIC- Cell Lines Project database so that it can be used by the global scientific community and clinicians to ultimately improve patient outcomes.
Dr A. Pia Sanzone, Non-Invasive Biomarkers and Whole Genome Analysis Group Leader at NIBSC comments that:
“Establishing this first series of WHO International Standards is only the beginning of a new era of development of WHO International Standards for somatic events. We will continue working - both to develop new standards and to further characterise the existing ones.”
Full details about our first pan-cancer genome standards can be found here. For more information on our International Standards for oncogene variant detection contact grmteam@nibsc.org.