With an ever-increasing number of clinically actionable genomic variants in cancer, there is an urgent need for the provision of standards to aid the definition of assay limit of detection and for the harmonisation of variant measurement in response to treatment. By using tumour cell lines carrying multiple rather than single actionable variants, the development of cancer genomic DNA (gDNA) standards may be accelerated.
In 2018, the WHO Expert Committee on Biological Standardization (ECBS) endorsed a new programme aimed at delivering a series of highly-characterised single-batch cancer gDNA (gDNA) International Standards covering multiple quantitative clinically-relevant variants. As additional variants within the WHO International Standards become clinically relevant, new data may be added, thus enhancing their clinical utility.
With our Cancer Genome WHO International Standards you will be able to:
- Define limits of detection or enable calibration and harmonisation of secondary standards, kits, and assays for the measurement of clinically-relevant variants using next-generation sequencing (NGS) and digital PCR (dPCR).
- Carry out the broader validation of any NGS pipeline, including long read sequencing, for other qualitative variants identified in the standards.
The standards are provided at the consensus variant percentage established from the international collaborative study and may be diluted using the corresponding wild-type WHO International Standard or an aligned wild-type gDNA, to generate further standards at a range of variant percentages which enable the calibration of quantitative assays. Additional variants identified in the standards during the collaborative study demonstrate suitability for the broader validation of any NGS pipeline.
Further information can be found on the product page for each standard:
Dilution to generate additional standards
The WHO International Standards for Cancer Genomes and the variant-carrying materials of the WHO 1st International Reference Panel for genomic KRAS codons 12 and 13 mutations may be diluted to produce further standards at lower variant percentages.
The preferable diluent for the WHO 1st International Standard for HCT 15 Cancer Genome (18/118), and the WHO 1st International Standard for MOLT-4 Cancer Genome (18/130) is the wild-type material WHO 1st International Standard for ATDB102 Reference Genome (18/164). The preferable diluent for the variant-carrying materials of the WHO 1st International Reference Panel for genomic KRAS codons 12 and 13 mutations is the KRAS wild-type panel member (material 16/266).
However, if insufficient wild-type standard is available to perform the dilutions, an alternative wild-type gDNA may be aligned to the wild-type standard and used as the diluent, i.e. it should be confirmed as being fully wild-type, diploid, and containing two copies of the gene of interest per diploid genome mass.
When preparing the dilutions, it is important to calculate the amount of wild-type gDNA needed to carry out all the dilutions required. A minimum of 5 dilution points (including the crude material) is recommended.
Dilutions of the nominal variants may be established as follows:
1) By use of the formula

where the variant copy number and total copy number per human diploid genome mass are specific for each variant and can be found in the Instructions for Use (IFU) document of the specific standard.
For example, to prepare a standard of 15% variant percentage for TP53 c.916C>T (R306*), the allelic content figures are used thus:
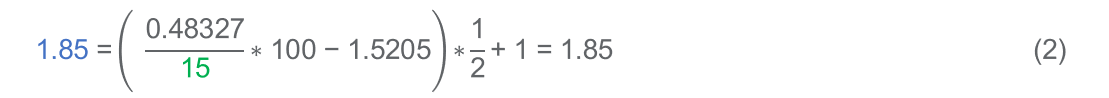
Meaning that a 1 in 1.85 dilution (in blue in example formula 2) of the 1st International Standard for MOLT-4 Cancer Genome (18/130) with the wild-type 1st International Standard for ATDB102 Reference Genome (18/164), will yield a further standard of consensus variant percentage 15% (in green in example formula 2) for TP53 c.916C>T (R306*), for example, 2.00 µl material 18/130, plus 1.70 µl material 18/164.
Notes:
- It is important to use the 5 decimal places for copy numbers in the calculation to achieve a maximally accurate answer.
- 0.48327 and 1.5205 in the above example are the are specific to TP53 c.916C>T (R306*) variant copy number and total copy number per human diploid genome mass in material 18/130 and can be found in Table 1 of the IFU. For each variant of interest please refer to the material’s specific IFU.
2) By reference to dilution curves available from NIBSC
An interactive dilution curve specific for each variant is provided on the product’s webpage.
For each variant, hover the “+” cursor over the dilution curve at the variant percentage required to see the dilution to be performed.
For example, to prepare a further standard of 15% variant percentage for TP53 c.916C>T (R306*) using the 1st International Standard for MOLT-4 Cancer Genome (18/130), hover the “+” cursor over 15% on the curve to see the dilution required i.e. 1.85 means that a 1 in 1.85 dilution (1 part of material 18/130 plus 0.85 parts of wild-type 1st International Standard for ATDB102 Reference Genome (18/164), will yield a further standard of variant percentage 15%, e.g.2.00 µl material 18/130, plus 1.70 µl material 18/164.
Notes:
- The links for the interactive dilution curves should be opened in Google Chrome. Performance in other browsers cannot be guaranteed.
- The variant percentage (%) is shown at 5 decimal places to ensure the accuracy of the dilution curves. Users are likely to be working with a maximum 1 or 2 decimal places so rounding may be required.
- For each variant of interest please refer to the interactive dilution curves on the specific product pages.
3) By use of pre-calculated dilutions
Refer to specific product Instructions for Use for details on the preparation of further standards for each nominal variant at a range of variant percentages.
How to order
To place an order please log in to your account or sign up for a new account.
For more information on our International Standards for oncogene variant detection contact grmteam@nibsc.org.